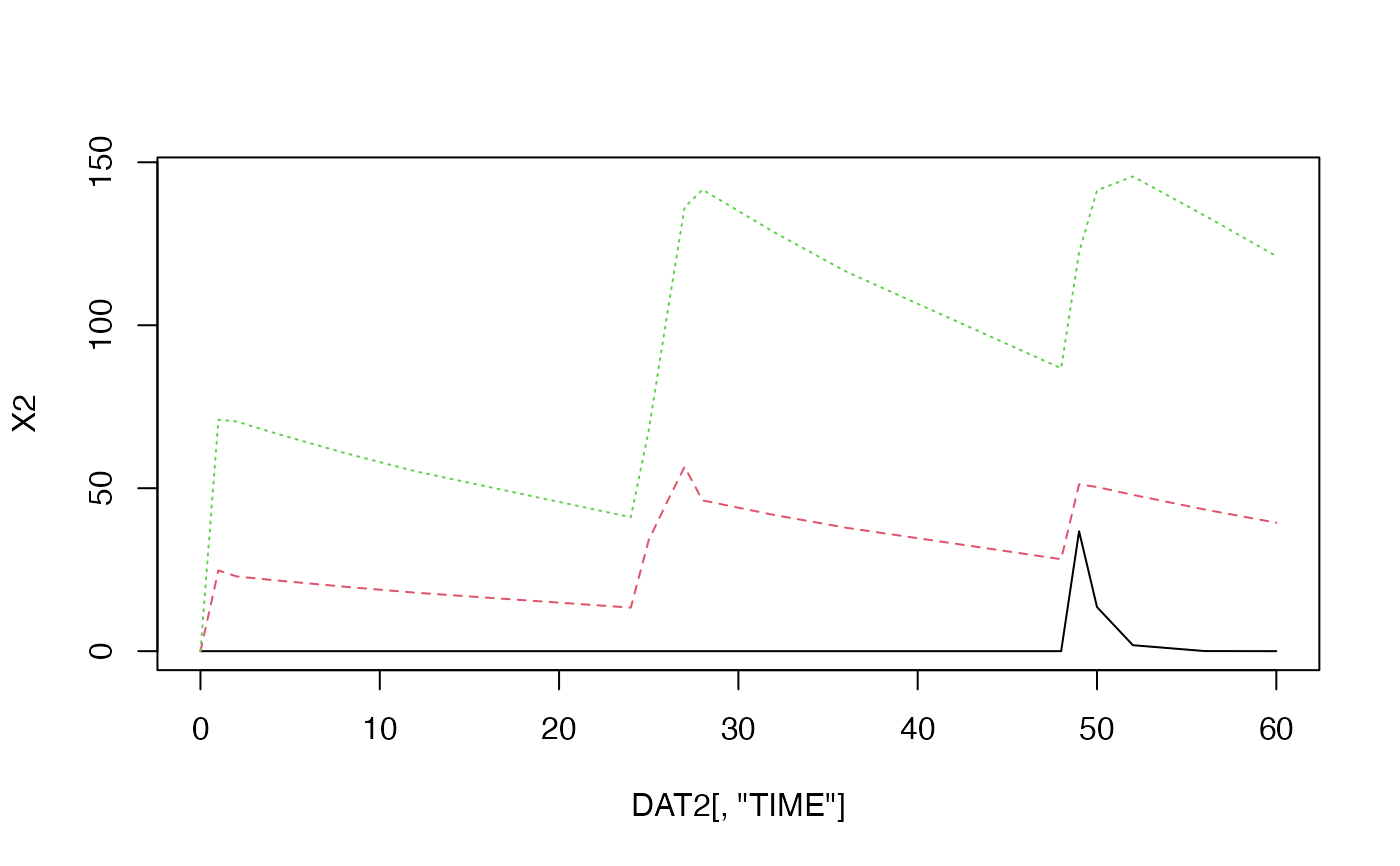
Get Amounts of Each Compartments using Lambdas and Coefficients of Multi-compartment Model
nComp.RdIt calculates using multi-compartment model.
nComp(Sol, Ka=0, DH)
Arguments
| Sol | Solution list of lambdas and coefficients |
|---|---|
| Ka | Absorption rate constant |
| DH | Expanded dosing history table |
Details
First compartment is the gut compartment for oral dosing. IV bolus and infusion dosing should be done at the second compartment. If a bolus dose was given at time T, it is reflected at times of larger than T. This is more close to real observation. ADAPT does like this, but NONMEM does not.
Value
This returns a table with the gut and the other compartment columns
Author
Kyun-Seop Bae <k@acr.kr>
Examples
DAT#> TIME AMT RATE CMT DV #> 1 0 100 0 2 NA #> 2 1 NA NA NA NA #> 3 2 NA NA NA NA #> 4 4 NA NA NA NA #> 5 8 NA NA NA NA #> 6 12 NA NA NA NA #> 7 24 150 50 2 NA #> 8 25 NA NA NA NA #> 9 26 NA NA NA NA #> 10 28 NA NA NA NA #> 11 32 NA NA NA NA #> 12 36 NA NA NA NA #> 13 48 100 0 1 NA #> 14 49 NA NA NA NA #> 15 50 NA NA NA NA #> 16 52 NA NA NA NA #> 17 56 NA NA NA NA #> 18 60 NA NA NA NA#> [,1] [,2] [,3] #> [1,] 0.000000e+00 0.00000 0.00000 #> [2,] 0.000000e+00 24.78579 71.00430 #> [3,] 0.000000e+00 22.94872 70.48922 #> [4,] 0.000000e+00 21.82887 67.13386 #> [5,] 0.000000e+00 19.78818 60.85782 #> [6,] 0.000000e+00 17.93827 55.16849 #> [7,] 0.000000e+00 13.36302 41.09748 #> [8,] 0.000000e+00 34.08869 67.74790 #> [9,] 0.000000e+00 45.53336 102.31559 #> [10,] 0.000000e+00 56.55153 136.19086 #> [11,] 0.000000e+00 46.25053 141.60095 #> [12,] 0.000000e+00 41.78336 128.50319 #> [13,] 0.000000e+00 37.87722 116.48999 #> [14,] 0.000000e+00 28.21644 86.77862 #> [15,] 3.678794e+01 51.20062 122.37609 #> [16,] 1.353353e+01 50.36158 141.38048 #> [17,] 1.831564e+00 47.95656 145.65708 #> [18,] 3.354626e-02 43.47332 133.66703 #> [19,] 6.144212e-04 39.40919 121.20089