One compartment model - analytical
Comp1.RdIt calculates using one compartment model.
Comp1(Ke, Ka=0, DH)
Arguments
| Ke | Elimination rate constant |
|---|---|
| Ka | Absorption rate constant |
| DH | Expanded dosing history table |
Details
First compartment is the gut compartment for oral dosing. IV bolus and infusion dosing should be done at the second compartment.
Value
This returns a table with the gut and the central compartment columns
Author
Kyun-Seop Bae <k@acr.kr>
Examples
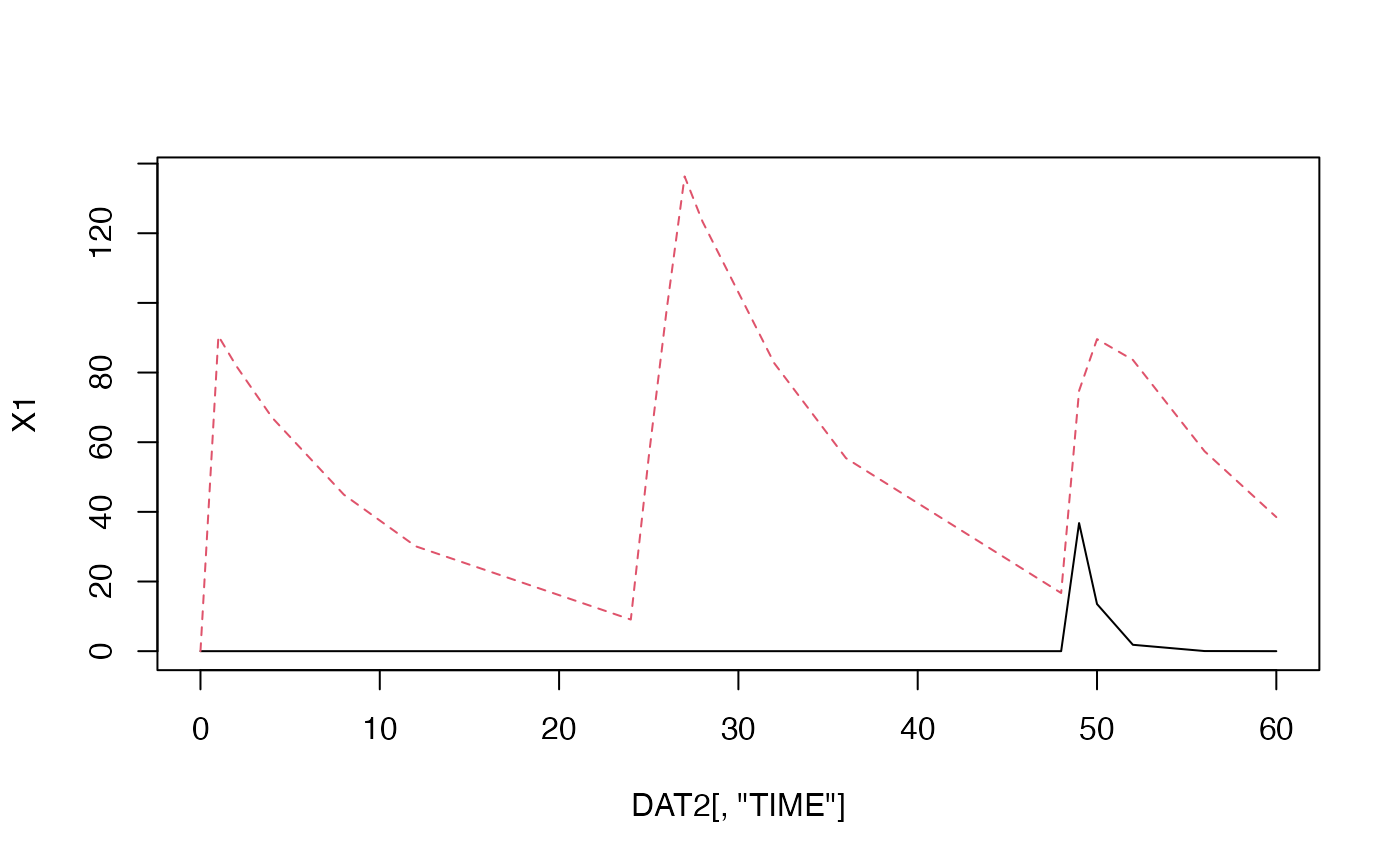
DAT#> TIME AMT RATE CMT DV #> 1 0 100 0 2 NA #> 2 1 NA NA NA NA #> 3 2 NA NA NA NA #> 4 4 NA NA NA NA #> 5 8 NA NA NA NA #> 6 12 NA NA NA NA #> 7 24 150 50 2 NA #> 8 25 NA NA NA NA #> 9 26 NA NA NA NA #> 10 28 NA NA NA NA #> 11 32 NA NA NA NA #> 12 36 NA NA NA NA #> 13 48 100 0 1 NA #> 14 49 NA NA NA NA #> 15 50 NA NA NA NA #> 16 52 NA NA NA NA #> 17 56 NA NA NA NA #> 18 60 NA NA NA NA#> [,1] [,2] #> [1,] 0.000000e+00 0.000000 #> [2,] 0.000000e+00 90.483742 #> [3,] 0.000000e+00 81.873075 #> [4,] 0.000000e+00 67.032005 #> [5,] 0.000000e+00 44.932896 #> [6,] 0.000000e+00 30.119421 #> [7,] 0.000000e+00 9.071795 #> [8,] 0.000000e+00 55.789791 #> [9,] 0.000000e+00 98.061981 #> [10,] 0.000000e+00 136.311441 #> [11,] 0.000000e+00 123.339692 #> [12,] 0.000000e+00 82.677068 #> [13,] 0.000000e+00 55.420096 #> [14,] 0.000000e+00 16.692212 #> [15,] 3.678794e+01 74.765736 #> [16,] 1.353353e+01 89.599257 #> [17,] 1.831564e+00 83.634059 #> [18,] 3.354626e-02 57.388461 #> [19,] 6.144212e-04 38.492939